-Search query
-Search result
Showing 1 - 50 of 56 items for (author: hwang & sh)

EMDB-40603: 
GPR161 Gs heterotrimer
Method: single particle / : Hoppe N, Manglik A, Harrison S

PDB-8smv: 
GPR161 Gs heterotrimer
Method: single particle / : Hoppe N, Manglik A, Harrison S

EMDB-35904: 
AtSLAC1 8D mutant in closed state
Method: single particle / : Lee Y, Lee S

EMDB-35920: 
AtSLAC1 in open state
Method: single particle / : Lee Y, Lee S

EMDB-34303: 
AtSLAC1 6D mutant in closed state
Method: single particle / : Lee Y, Lee S

EMDB-34304: 
AtSLAC1 6D mutant in open state
Method: single particle / : Lee Y, Lee S

EMDB-29013: 
96nm doublet microtubule repeat from wild type mouse sperm
Method: subtomogram averaging / : Hwang JY, Chai P, Nawaz S

EMDB-28606: 
96nm doublet microtubule repeat from LRRC23-KO mouse sperm
Method: subtomogram averaging / : Hwang JY, Chai P, Nawaz S

EMDB-34087: 
The cargo delivery vehicle Hcp-VgrG-PAAR of the Type VI secretion system
Method: single particle / : He W, Wu K, Zhu L, Wen Y

PDB-8gra: 
Structure of Type VI secretion system cargo delivery vehicle Hcp-VgrG-PAAR
Method: single particle / : Wen Y, He W, Zhu L

EMDB-33734: 
Cryo-EM structure of SARS-CoV-2 spike in complex with K202.B bispecific antibody
Method: single particle / : Yoo Y, Cho HS

EMDB-25427: 
Structure of KRAS G12V/HLA-A*03:01 in complex with antibody fragment V2
Method: single particle / : Wright KM, Gabelli SB

PDB-7stf: 
Structure of KRAS G12V/HLA-A*03:01 in complex with antibody fragment V2
Method: single particle / : Wright KM, Gabelli SB, Miller M

EMDB-26989: 
Extracellular architecture of an engineered canonical Wnt signaling ternary complex
Method: single particle / : Tsutsumi N, Jude KM, Garcia KC

PDB-8ctg: 
Extracellular architecture of an engineered canonical Wnt signaling ternary complex
Method: single particle / : Tsutsumi N, Jude KM, Garcia KC
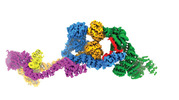
EMDB-29073: 
Human IFT-A complex structures provide molecular insights into ciliary transport
Method: single particle / : Jiang M, Palicharla VR, Miller D, Hwang SH, Zhu H, Hixson P, Mukhopadhyay S, Sun J
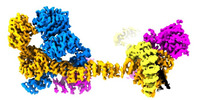
EMDB-29078: 
Human IFT-A complex structures provide molecular insights into ciliary transport
Method: single particle / : Jiang M, Palicharla VR, Miller D, Hwang SH, Zhu H, Hixson P, Mukhopadhyay S, Sun J
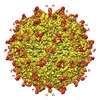
EMDB-25606: 
Zika Virus particle bound with IgM antibody DH1017 Fab fragment
Method: single particle / : Miller AS, Kuhn RJ

PDB-7t17: 
Zika Virus asymmetric unit bound with IgM antibody DH1017 Fab fragment
Method: single particle / : Miller AS, Kuhn RJ

EMDB-33466: 
Cryo-EM structure of HK022 putRNA-associated E.coli RNA polymerase elongation complex
Method: single particle / : Hwang SH, Kang JY

EMDB-24210: 
The 3D structure and in situ arrangements of CatSper channel from the cryo-electron tomography and subtomographic average of mouse wild type sperm flagella
Method: subtomogram averaging / : Zhao Y, Wang H, Wiesehoefer C, Shah NB, Reetz E, Hwang J, Huang X, Lishko PV, Davies KM, Wennemuth G, Nicastro D, Chung JJ

EMDB-26206: 
The 3D structure and in situ arrangements (Forward slash) of CatSper channel from the cryo-electron tomography and subtomographic average of mouse efcab9 mutant sperm flagella
Method: subtomogram averaging / : Zhao Y, Wang H, Wiesehoefer C, Shah NB, Reetz E, Hwang J, Huang X, Lishko PV, Davies KM, Wennemuth G, Nicastro D, Chung JJ

EMDB-26207: 
The 3D structure and in situ arrangements (Backslash) of CatSper channel from the cryo-electron tomography and subtomographic average of mouse efcab9 mutant sperm flagella
Method: subtomogram averaging / : Zhao Y, Wang H, Wiesehoefer C, Shah NB, Reetz E, Hwang J, Huang X, Lishko PV, Davies KM, Wennemuth G, Nicastro D, Chung JJ

EMDB-30673: 
Activity optimized supercomplex state1
Method: single particle / : Jeon TJ, Lee SG, Yoo SH, Ryu JH, Kim DS, Hyun JK, Kim HM, Ryu SE

EMDB-30674: 
Activity optimized supercomplex state2
Method: single particle / : Jeon TJ, Lee SG, Yoo SH, Ryu JH, Kim DS, Hyun JK, Kim HM, Ryu SE

EMDB-30675: 
Activity optimized supercomplex state3
Method: single particle / : Jeon TJ, Lee SG, Yoo SH, Ryu JH, Kim DS, Hyun JK, Kim HM, Ryu SE

EMDB-30676: 
Activity optimized complex I (closed form)
Method: single particle / : Jeon TJ, Lee SG, Yoo SH, Ryu JH, Kim DS, Hyun JK, Kim HM, Ryu SE

EMDB-30677: 
Activity optimized complex I (open form)
Method: single particle / : Jeon TJ, Lee SG, Yoo SH, Ryu JH, Kim DS, Hyun JK, Kim HM, Ryu SE

EMDB-30706: 
Activity optimized supercomplex state4
Method: single particle / : Jeon TJ, Lee SG, Yoo SH, Ryu JH, Kim DS, Hyun JK, Kim HM, Ryu SE

EMDB-31470: 
Cryo-EM structure of SARS-CoV-2 spike in complex with a neutralizing antibody chAb-25 (Focused refinement of S-RBD and chAb-25 region)
Method: single particle / : Yang TJ, Yu PY, Wu HC, Hsu STD

EMDB-31471: 
Cryo-EM structure of SARS-CoV-2 spike in complex with a neutralizing antibody chAb-45 (Focused refinement of S-RBD and chAb-45 region)
Method: single particle / : Yang TJ, Yu PY, Wu HC, Hsu STD

PDB-7f62: 
Cryo-EM structure of SARS-CoV-2 spike in complex with a neutralizing antibody chAb-25 (Focused refinement of S-RBD and chAb-25 region)
Method: single particle / : Yang TJ, Yu PY, Wu HC, Hsu STD

PDB-7f63: 
Cryo-EM structure of SARS-CoV-2 spike in complex with a neutralizing antibody chAb-45 (Focused refinement of S-RBD and chAb-45 region)
Method: single particle / : Yang TJ, Yu PY, Wu HC, Hsu STD

EMDB-30929: 
The negatively stained reconstruction of cytoplasmic AdeS in APO state
Method: single particle / : Zhu L, Wu D, Wu K

EMDB-30930: 
The negatively stained reconstruction of cytoplasmic AdeS in ATP-bound state
Method: single particle / : Zhu L, Wu D, Wu K

EMDB-12303: 
Mammalian ribosome nascent chain complex with SRP and SRP receptor in early state A
Method: single particle / : Jomaa A, Lee JH, Shan S, Ban N

EMDB-12304: 
SRP54 and SRP RNA proximal site
Method: single particle / : Jomaa A, Ban N

EMDB-12305: 
Mammalian ribosome nascent chain complex with SRP and SRP receptor in the early state B
Method: single particle / : Jomaa A, Lee JH, Shan S, Ban N

PDB-7nfx: 
Mammalian ribosome nascent chain complex with SRP and SRP receptor in early state A
Method: single particle / : Jomaa A, Lee JH, Shan S, Ban N

EMDB-23156: 
SARS-CoV 2 Spike Protein bound to LY-CoV555
Method: single particle / : Goldsmith JA, McLellan JS

PDB-7l3n: 
SARS-CoV 2 Spike Protein bound to LY-CoV555
Method: single particle / : Goldsmith JA, McLellan JS

EMDB-22295: 
Cryo-EM structure of SHIV-elicited RHA1.V2.01 in complex with HIV-1 Env BG505 DS-SOSIP.664
Method: single particle / : Gorman J, Kwong PD

PDB-6xrt: 
Cryo-EM structure of SHIV-elicited RHA1.V2.01 in complex with HIV-1 Env BG505 DS-SOSIP.664
Method: single particle / : Gorman J, Kwong PD

EMDB-11002: 
CryoEM structure of bovine cytochrome bc1 in complex with a tetrahydroquinolone inhibitor
Method: single particle / : Muench SP, Johnson R, Amporndanai K, Atonyuk S

EMDB-3779: 
Cryo-electron tomography averaged map of microtubule doublet from p23 mutant Chlamydomonas axoneme
Method: subtomogram averaging / : Sale WS, Ishikawa T, Yamamoto R, Obbineni JM, Alford LM, Hwang J, Ide T, Owa M, Kon T, Inaba K, Noliyanda J, King SM, Dutcher S

EMDB-3786: 
Cryo-electron tomography averaged map of microtubule doublet from p23 mutant Chlamydomonas axoneme
Method: subtomogram averaging / : Sale WS, Ishikawa T, Yamamoto R, Obbineni JM, Alford LM, Hwang J, Ide T, Owa M, Kon T, Inaba K, Noliyanda J, King SM, Dutcher S

EMDB-3787: 
Cryo-electron tomography averaged map of microtubule doublet from p23 mutant Chlamydomonas axoneme
Method: subtomogram averaging / : Sale WS, Ishikawa T, Yamamoto R, Obbineni JM, Alford LM, Hwang J, Ide T, Owa M, Kon T, Inaba K, Noliyanda J, King SM, Dutcher S

PDB-5nco: 
Quaternary complex between SRP, SR, and SecYEG bound to the translating ribosome
Method: single particle / : Jomaa A, Hwang Fu Y, Boerhinger D, Leibundgut M, Shan SO, Ban N
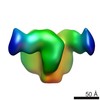
EMDB-8507: 
92BR SOSIP.664 trimer in complex with DH270.1 Fab
Method: single particle / : Fera D, Harrison SC
EMDB-8078: 
BG505 SOSIP.664 HIV-1 Env trimer in complex with anti-HIV CH235.12 wk323 Fab
Method: single particle / : Ozorowski G, Ward AB
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
